How to boost gene expression?
synthetic biology, transgene, solubility
Why are some genes highly expressed? Why are some proteins poorly soluble? We sought to understand the key sequence features that control gene expression and protein solubility. We identify that the ‘accessibility’ of the mRNA regions around initiation codons correlates with gene expression (Bhandari et al., 2021). We find that ‘flexible’ proteins tend to be more soluble (Bhandari et al., 2020). We have developed user-friendly web services to harness these features for cost-effective designs of mRNAs and proteins (Bhandari et al., 2021).
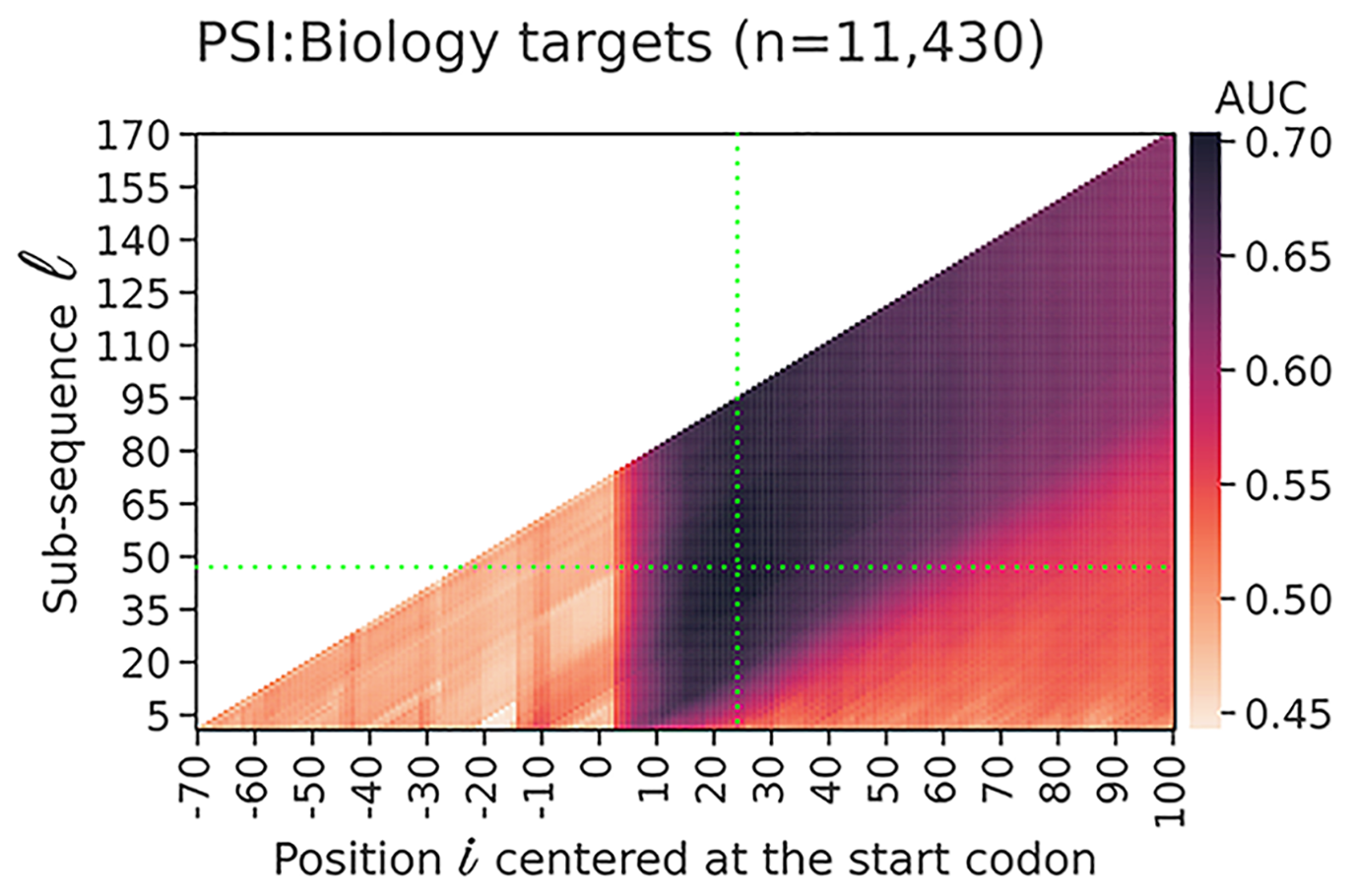
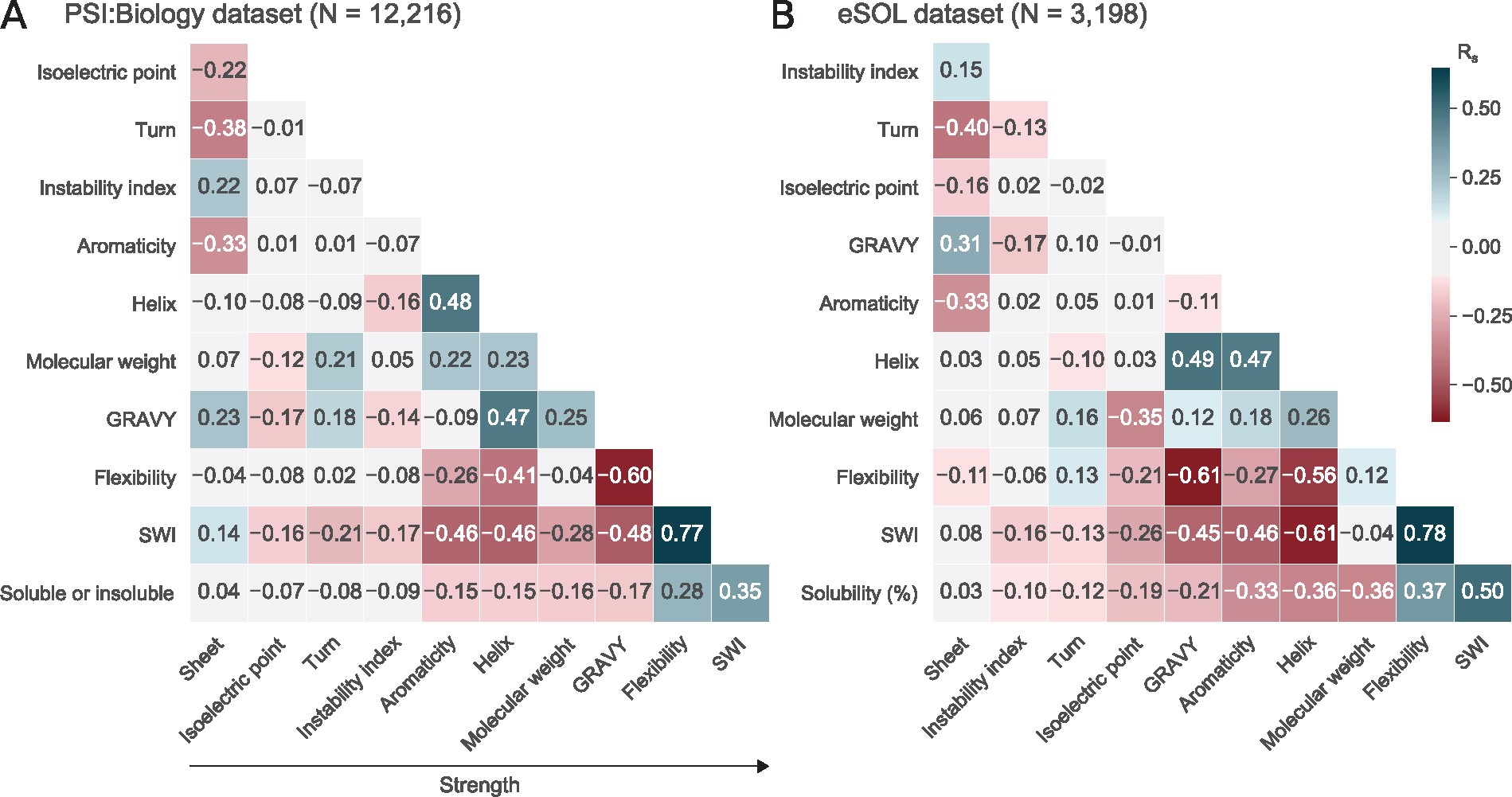
Accessibility around start codons are predictive of protein expression in E. coli (left). Highly flexible proteins tend to be more soluble (right). Based on this, we created a new index called Solubility-Weighted Index to predict solubility.
References
2021
- Analysis of 11,430 recombinant protein production experiments reveals that protein yield is tunable by synonymous codon changes of translation initiation sitesPLOS Computational Biology, 2021
- TISIGNER.com: web services for improving recombinant protein productionNucleic Acids Research, 2021
2020
- Solubility-Weighted Index: fast and accurate prediction of protein solubilityBioinformatics, 2020